Imposing an interaface condition in Poisson problem, serial version. More...
#include <poisson_nitsche_interface.h>
Public Member Functions | |
| PoissonNitscheInterface () | |
| void | run () |
 Public Member Functions inherited from ParameterAcceptor Public Member Functions inherited from ParameterAcceptor | |
| ParameterAcceptor (const std::string §ion_name="") | |
| unsigned int | get_acceptor_id () const |
| virtual | ~ParameterAcceptor () override |
| virtual void | declare_parameters (ParameterHandler &prm) |
| virtual void | parse_parameters (ParameterHandler &prm) |
| std::string | get_section_name () const |
| std::vector< std::string > | get_section_path () const |
| void | add_parameter (const std::string &entry, ParameterType ¶meter, const std::string &documentation="", ParameterHandler &prm_=prm, const Patterns::PatternBase &pattern=*Patterns::Tools::Convert< ParameterType >::to_pattern()) |
| void | enter_subsection (const std::string &subsection) |
| void | leave_subsection () |
| void | enter_my_subsection (ParameterHandler &prm) |
| void | leave_my_subsection (ParameterHandler &prm) |
| void | serialize (Archive &ar, const unsigned int version) |
| void | subscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| void | unsubscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| unsigned int | n_subscriptions () const |
| void | list_subscribers (StreamType &stream) const |
| void | list_subscribers () const |
| void | subscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| void | unsubscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| unsigned int | n_subscriptions () const |
| void | list_subscribers (StreamType &stream) const |
| void | list_subscribers () const |
 Public Member Functions inherited from Subscriptor Public Member Functions inherited from Subscriptor | |
| Subscriptor () | |
| Subscriptor (const Subscriptor &) | |
| Subscriptor (Subscriptor &&) noexcept | |
| virtual | ~Subscriptor () |
| Subscriptor & | operator= (const Subscriptor &) |
| Subscriptor & | operator= (Subscriptor &&) noexcept |
| void | serialize (Archive &ar, const unsigned int version) |
| void | subscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| void | unsubscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| unsigned int | n_subscriptions () const |
| void | list_subscribers (StreamType &stream) const |
| void | list_subscribers () const |
| void | subscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| void | unsubscribe (std::atomic< bool > *const validity, const std::string &identifier="") const |
| unsigned int | n_subscriptions () const |
| void | list_subscribers (StreamType &stream) const |
| void | list_subscribers () const |
Protected Member Functions | |
| void | generate_grids () |
| void | setup_system () |
| Setup dofs, constraints, and matrices. More... | |
| void | assemble_system () |
| void | solve () |
| void | output_results (const unsigned cycle) const |
Protected Attributes | |
| const std::string | component_names = "u" |
| ParsedTools::GridGenerator< spacedim, spacedim > | grid_generator |
| ParsedTools::GridGenerator< dim, spacedim > | embedded_grid_generator |
| ParsedTools::GridRefinement | grid_refinement |
| Triangulation< spacedim, spacedim > | space_triangulation |
| The actual triangulations. More... | |
| Triangulation< dim, spacedim > | embedded_triangulation |
| std::unique_ptr< GridTools::Cache< spacedim, spacedim > > | space_cache |
| GridTools::Cache objects are used to cache all the necessary information about a given triangulation, such as its Mapping, Bounding Boxes, etc. More... | |
| std::unique_ptr< GridTools::Cache< dim, spacedim > > | embedded_cache |
| std::vector< std::tuple< typename Triangulation< spacedim, spacedim >::cell_iterator, typename Triangulation< dim, spacedim >::cell_iterator, Quadrature< spacedim > > > | cells_and_quads |
| The coupling between the two grids is ultimately encoded in this vector. More... | |
| ParsedTools::FiniteElement< spacedim, spacedim > | space_fe |
| DoFHandler< spacedim, spacedim > | space_dh |
| The actual DoFHandler class. More... | |
| std::unique_ptr< Mapping< spacedim, spacedim > > | mapping |
| According to the Triangulation type, we use a MappingFE or a MappingQ, to make sure we can run the program both on a tria/tetra grid and on quad/hex grids. More... | |
Linear algebra classes | |
| AffineConstraints< double > | space_constraints |
| SparsityPattern | sparsity_pattern |
| SparseMatrix< double > | system_matrix |
| Vector< double > | solution |
| Vector< double > | system_rhs |
| ParsedLAC::InverseOperator | inverse_operator |
| ParsedLAC::AMGPreconditioner | preconditioner |
Forcing terms and boundary conditions | |
| ParsedTools::Constants | constants |
| ParsedTools::Function< spacedim > | forcing_term |
| The actual function to use as a forcing term. More... | |
| ParsedTools::Function< spacedim > | embedded_value |
| This is the value we want to impose on the embedded domain. More... | |
| ParsedTools::Function< spacedim > | nitsche_coefficient |
| The coefficient in front of the Nitsche contribution to the stiffness matrix. More... | |
| ParsedTools::Function< spacedim > | exact_solution |
| The actual function to use as a exact solution when computing the errors. More... | |
| ParsedTools::BoundaryConditions< spacedim > | boundary_conditions |
Output and postprocessing | |
| TimerOutput | timer |
| ParsedTools::ConvergenceTable | error_table |
| ParsedTools::DataOut< spacedim, spacedim > | data_out |
| Choosing as embedded space the square \([-.0.45,0.45]^2\) and as embedding space the square \([-1,1]^2\), with embedded value the function \(g(x,y)=1\), this is what we get. More... | |
| unsigned int | console_level = 1 |
| Level of log verbosity. More... | |
| double | penalty = 100.0 |
| The penalty parameter which multiplies Nitsche's terms. More... | |
 Protected Attributes inherited from ParameterAcceptor Protected Attributes inherited from ParameterAcceptor | |
| const std::string | section_name |
| std::vector< std::string > | subsections |
Additional Inherited Members | |
 Static Public Member Functions inherited from ParameterAcceptor Static Public Member Functions inherited from ParameterAcceptor | |
| static void | initialize (const std::string &filename="", const std::string &output_filename="", const ParameterHandler::OutputStyle output_style_for_output_filename=ParameterHandler::Short, ParameterHandler &prm=ParameterAcceptor::prm, const ParameterHandler::OutputStyle output_style_for_filename=ParameterHandler::DefaultStyle) |
| static void | initialize (std::istream &input_stream, ParameterHandler &prm=ParameterAcceptor::prm) |
| static void | clear () |
| static void | parse_all_parameters (ParameterHandler &prm=ParameterAcceptor::prm) |
| static void | declare_all_parameters (ParameterHandler &prm=ParameterAcceptor::prm) |
| static ::ExceptionBase & | ExcInUse (int arg1, std::string arg2, std::string arg3) |
| static ::ExceptionBase & | ExcNoSubscriber (std::string arg1, std::string arg2) |
 Static Public Member Functions inherited from Subscriptor Static Public Member Functions inherited from Subscriptor | |
| static ::ExceptionBase & | ExcInUse (int arg1, std::string arg2, std::string arg3) |
| static ::ExceptionBase & | ExcNoSubscriber (std::string arg1, std::string arg2) |
 Public Attributes inherited from ParameterAcceptor Public Attributes inherited from ParameterAcceptor | |
| boost::signals2::signal< void()> | declare_parameters_call_back |
| boost::signals2::signal< void()> | parse_parameters_call_back |
 Static Public Attributes inherited from ParameterAcceptor Static Public Attributes inherited from ParameterAcceptor | |
| static ParameterHandler | prm |
Imposing an interaface condition in Poisson problem, serial version.
Here we solve the Poisson equation:
\[ \begin{cases} - \Delta u = f & \text{ in } \Omega \subset R^{\text{spacedim}}\\ u = u_D = 0 & \text{ on } \partial \Omega \\ u = g & \text{on} B \\ \end{cases} \]
where B is a domain embedded in \Omega. The structure of the program is the usual one, what really changes is the weak form. To get that, we apply Nitsche's method in \(\Omega \setminus B\) and \(B\), separately, as done in step-70 tutorial. For an extended discussion see there and references therein. All in all, what one gets using Lagrangian finite elements in codimension 1 is to find \(u \in H_0^1(\Omega)\) s.t.
\begin{align*} (\nabla u, \nabla v) + 2 \beta \langle u,v \rangle_{\Gamma} =(f,v) + 2 \beta \langle g,v \rangle_{\Gamma} \end{align*}
for all test functions \(v \in H_0^1(\Omega)\). Here \(\Gamma = \partial B\) in codimension 1. The contributions from Nitsche's method then have to be added both to the stiffness matrix and to the right hand side. The thing here is that they have to be computed on the embedded grid, while \(u\) and \(v\) live on the ambient space.
Definition at line 107 of file poisson_nitsche_interface.h.
| PDEs::Serial::PoissonNitscheInterface< dim, spacedim >::PoissonNitscheInterface |
Definition at line 31 of file poisson_nitsche_interface.cc.
| void PDEs::Serial::PoissonNitscheInterface< dim, spacedim >::run |
Definition at line 282 of file poisson_nitsche_interface.cc.
References deallog, LogStream::depth_console(), Utilities::int_to_string(), LogStream::pop(), and LogStream::push().
|
protected |
Definition at line 70 of file poisson_nitsche_interface.cc.
|
protected |
Setup dofs, constraints, and matrices.
This function enumerate all the degrees of freedom and set up matrix and vector objects to hold the system data. Enumerating is done by using DoFHandler::distribute_dofs(), as we have seen in the step-2 example, with the difference that now we pass a ParsedTools::FiniteElement object instead of directly a FiniteElement object.
Definition at line 88 of file poisson_nitsche_interface.cc.
References AssertThrow, deallog, StandardExceptions::ExcMessage(), get_default_linear_mapping(), DoFTools::make_hanging_node_constraints(), DoFTools::make_sparsity_pattern(), and reference_cell().
|
protected |
Definition at line 141 of file poisson_nitsche_interface.cc.
References deallog, ReferenceCell::get_gauss_type_quadrature(), update_gradients, update_JxW_values, update_quadrature_points, and update_values.
|
protected |
Definition at line 238 of file poisson_nitsche_interface.cc.
References deallog, and inverse_operator().
|
protected |
Definition at line 255 of file poisson_nitsche_interface.cc.
References deallog, Utilities::int_to_string(), Utilities::needed_digits(), and GridOut::write_vtk().
|
protected |
Definition at line 148 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 151 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 153 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 155 of file poisson_nitsche_interface.h.
|
protected |
The actual triangulations.
Here with "space_triangulation" we refer to the original domain \Omega, also called the ambient space, while with embedded we refer to the immersed domain, the one where we want to impose a constraint.
Definition at line 164 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 165 of file poisson_nitsche_interface.h.
|
protected |
GridTools::Cache objects are used to cache all the necessary information about a given triangulation, such as its Mapping, Bounding Boxes, etc.
Definition at line 173 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 174 of file poisson_nitsche_interface.h.
|
protected |
The coupling between the two grids is ultimately encoded in this vector.
Here the i-th entry stores a tuple for which the first two elements are iterators to two cells from the space and embedded grid, respectively, that intersect each other (up to a specified tolerance) and a Quadrature object to integrate over that region.
Definition at line 189 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 192 of file poisson_nitsche_interface.h.
|
protected |
The actual DoFHandler class.
Definition at line 197 of file poisson_nitsche_interface.h.
|
protected |
According to the Triangulation type, we use a MappingFE or a MappingQ, to make sure we can run the program both on a tria/tetra grid and on quad/hex grids.
Definition at line 204 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 211 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 212 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 213 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 214 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 215 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 216 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 217 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 226 of file poisson_nitsche_interface.h.
|
protected |
The actual function to use as a forcing term.
Definition at line 231 of file poisson_nitsche_interface.h.
|
protected |
This is the value we want to impose on the embedded domain.
Definition at line 238 of file poisson_nitsche_interface.h.
|
protected |
The coefficient in front of the Nitsche contribution to the stiffness matrix.
Definition at line 246 of file poisson_nitsche_interface.h.
|
protected |
The actual function to use as a exact solution when computing the errors.
Definition at line 252 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 255 of file poisson_nitsche_interface.h.
|
mutableprotected |
Definition at line 262 of file poisson_nitsche_interface.h.
|
protected |
Definition at line 265 of file poisson_nitsche_interface.h.
|
mutableprotected |
Choosing as embedded space the square \([-.0.45,0.45]^2\) and as embedding space the square \([-1,1]^2\), with embedded value the function \(g(x,y)=1\), this is what we get.
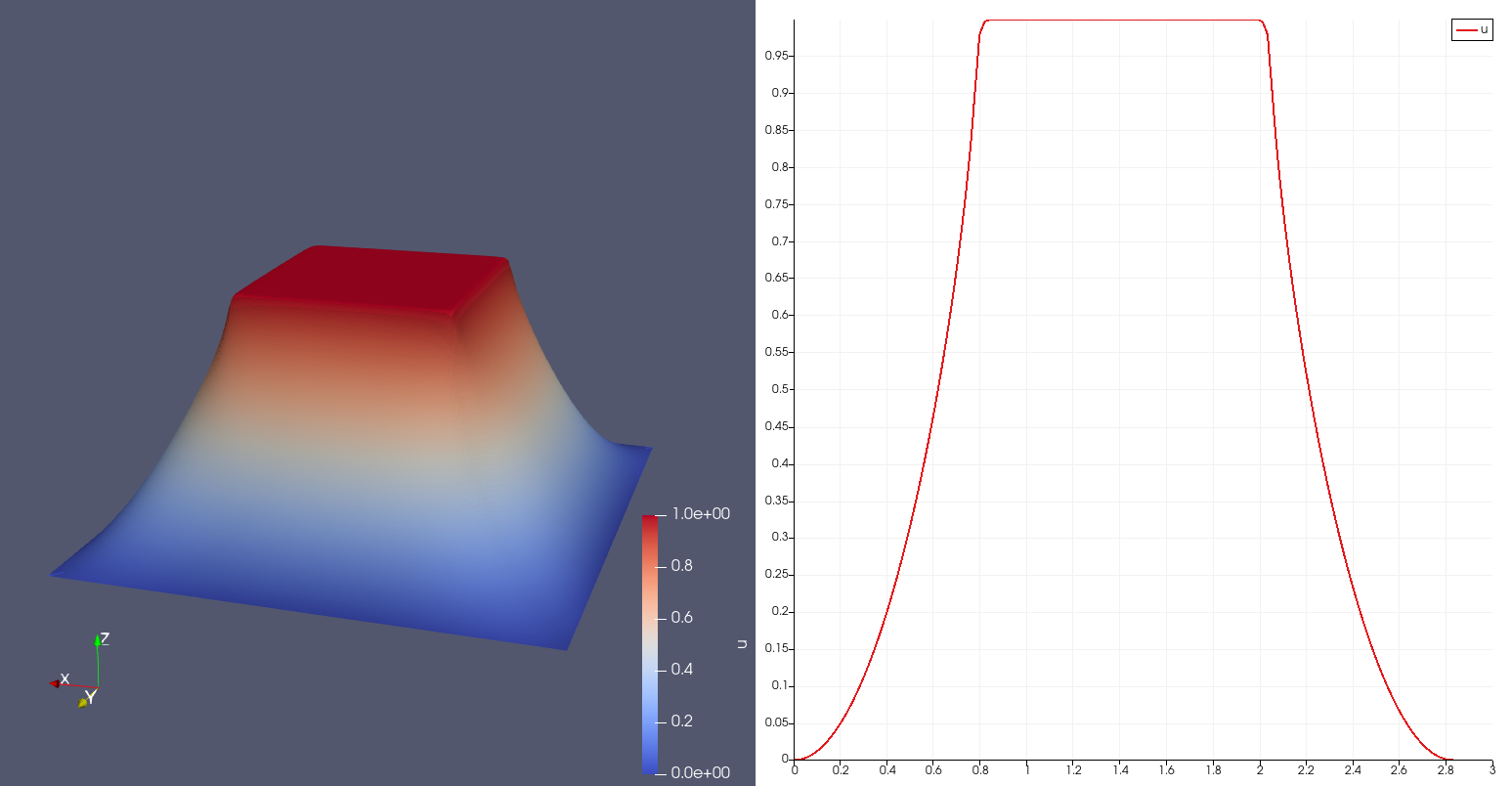
Taking a manufactured smooth solution \(u=\sin(2 \pi x) \sin(2 \pi y)\), classical rates can be observed, as in the following table: cells dofs u_L2_norm u_Linfty_norm u_H1_norm 256 289 5.851e-02 - 8.125e-02 - 2.015e+00 - 1024 1089 1.436e-02 2.12 2.160e-02 2.00 1.007e+00 1.05 4096 4225 3.605e-03 2.04 5.519e-03 2.01 5.037e-01 1.02
Definition at line 283 of file poisson_nitsche_interface.h.
|
protected |
Level of log verbosity.
Definition at line 289 of file poisson_nitsche_interface.h.
|
protected |
The penalty parameter which multiplies Nitsche's terms.
In this program it is defaulted to 100.0
Definition at line 299 of file poisson_nitsche_interface.h.